A complete analysis of the microbiome should combine taxonomic profiling with functional characterization to discover the full potential of an entire community of microorganisms. However, research teams often require specialized training and extensive computational infrastructure to establish an in-house suite of tools for processing shotgun metagenomic data – and even with the right know-how, the processing can be both time-consuming and challenging. That’s why, in the past year, CosmosID-HUB introduced a new functional profiling workflow that allows users to get community-level MetaCyc Pathways and GO Terms alongside its patented strain level taxonomic profiling technology. The CosmosID-HUB makes it easy and intuitive to analyze microbiome taxonomic and functional data without the need for in-depth knowledge in bioinformatics.
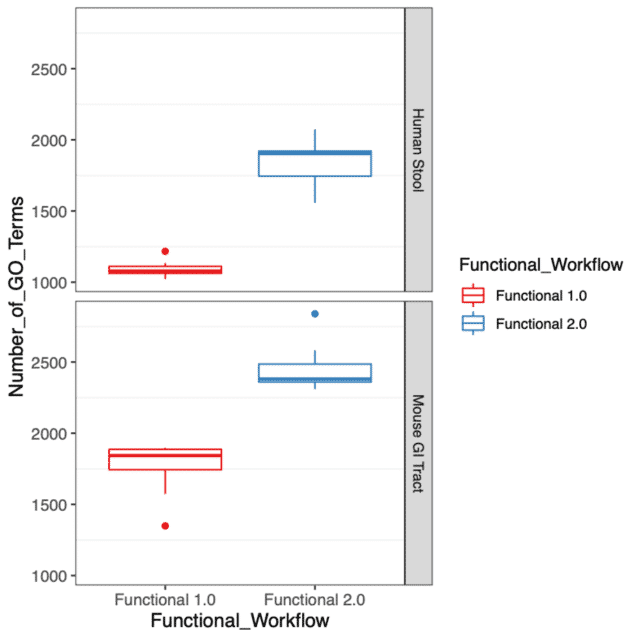
This year, we are pleased to announce two major updates to our functional profiling workflow. One of the primary focuses of this latest release was to improve the functional characterization of the microbiome by expanding our catalog of gene families from Uniref90. The update to the Uniref90 gene family database has substantially increased our detection of gene functions in the microbiome community, which can be seen in Figure 1 with an increase in the number of GO Terms detected in both human stool and mouse gastrointestinal tract specimens.
Furthermore, alongside MetaCyc Pathways and GO Terms database, CosmosID-HUB users will have access to 3 more additional functional databases as well. The three additional databases are:
1) Enzyme Commission Database: a database of the numerical classification of enzymes based on the reactions they catalyze
2) Pfam Database: a database of protein families generated using hidden Markov models
3) CAZy Database: a database of structurally related catalytic and carbohydrate-binding modules of enzymes that degrade, modify, or create glycosidic bonds
The addition of these three new databases will give scientists greater insights into the functional potential of their microbial communities at the gene/protein level within minutes of uploading their raw FASTQ files to the HUB.
The release of the Functional Workflow 2.0 is part of CosmosID’s microbial genomics platform, CosmosID-HUB Microbiome, which is continually expanding with various features and workflows for analyzing microbial communities and for inferring translational and actionable microbiome insights.
Currently, CosmosID-HUB Microbiome analyzes 16S, whole genome (shotgun metagenomics and metatranscriptomics), and ITS sequencing data and provides options for discovering antimicrobial resistance (AMR) and virulence markers, microbial taxa (composition), and functions.
Users are also able to aggregate and compare their primary analysis results (taxa, AMR and virulence markers, functions) by cohorts or groups using statistical modules to gain actionable insights from their data.


