The Kepler multi-kingdom taxonomic profiler is divided into three parts:
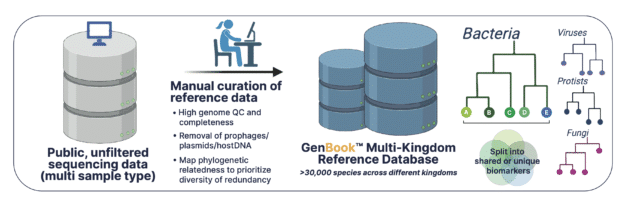
The Kepler database of high quality microbial genomes is based on high completeness:low contamination ratio, genome assembly quality and prioritizing intra-species diversity whilst limiting phylogenetic redundancy. The genome assemblies are then scrubbed clean of low complexity sequences, prophages, plasmids and host-contaminated regions to maximize the taxonomic signal-to-noise ratio. The final database encompasses multiple microbial kingdoms and >30,000 species.
2. Identifying Relevant Biomarkers
With the genomes curated and cleaned, they undergo a pre-computation phase where they’re split into n-mers of variable length. The n-mers are then categorized as either shared or unique biomarkers across individual genomes, which is facilitated by a phylogenetic tree-like data structure. The tree backbone represents shared genomic biomarkers between different taxa, while the tree leaves are individual microbial genomes with unique biomarkers.
3. Searching the Biomarker Database
The second per-sample, computational phase searches the millions of short sequence reads or contigs in your data against the phylogenetic tree-like database build:
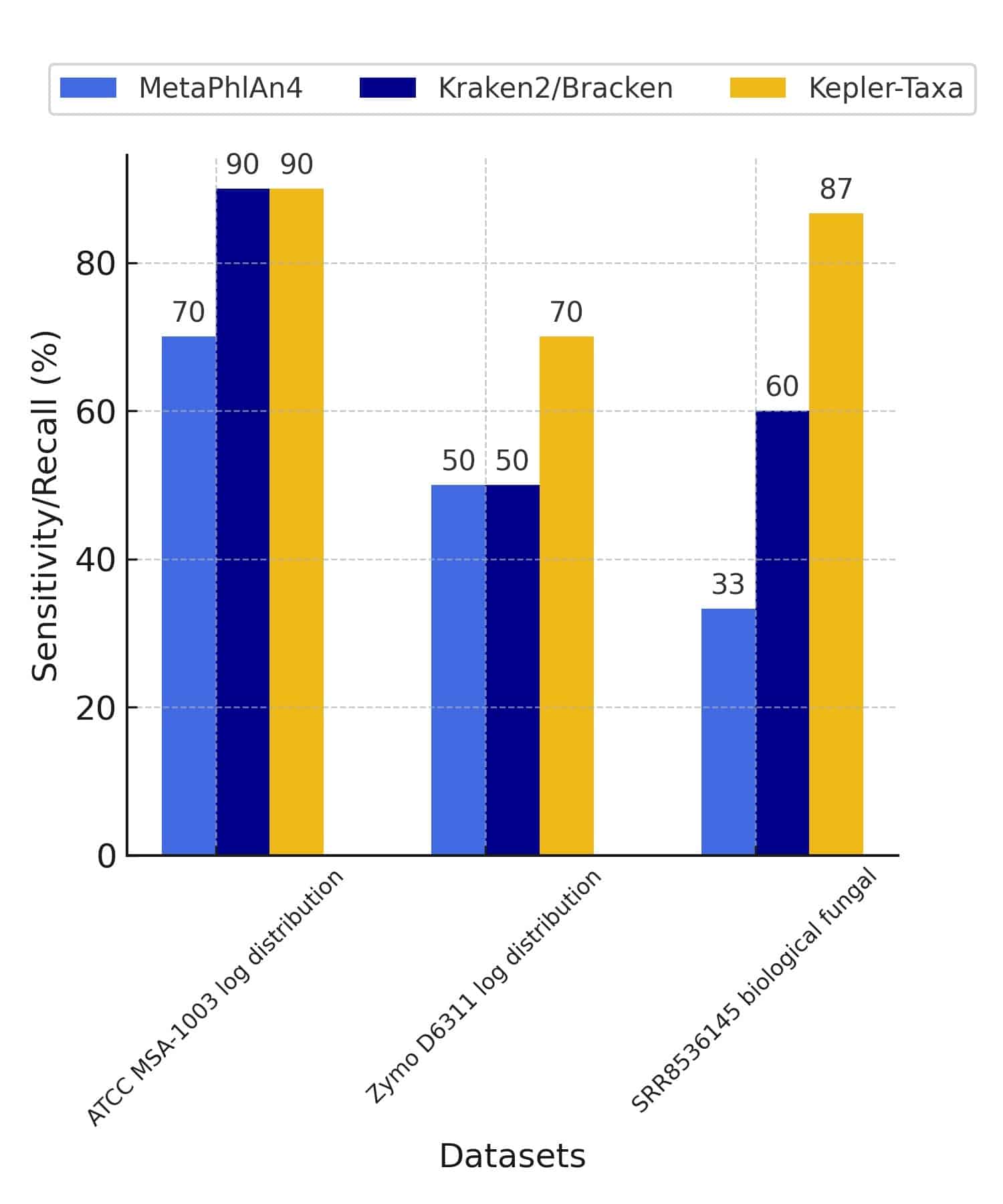
A. The first comparator splits the sequencing reads into k-mer sets that are then queried across the different branches and leaves of the phylogenetic tree to identify the different taxa present in the query kmer-sets. The first comparator splits the sequencing reads into k-mer sets that are then queried across the different branches and leaves of the phylogenetic tree to identify the different taxa present in the query kmer-sets. The first comparator looks for exact matches between query k-mers and reference bio-markers and classification sensitivity and accuracy is maintained through composite k-mer/biomarker aggregation statistics and coverage depth estimation.
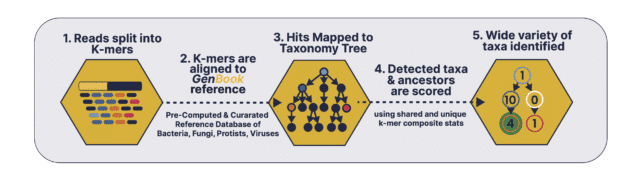
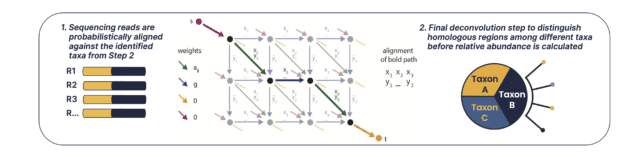
B. The second comparator uses an edit distance-scoring based probabilistic Smith-Waterman algorithm to compare sequencing reads with a reference set of identified microbial taxa using the first comparator. In conclusion, overall abundance precision and classification accuracy is achieved by running the comparators in sequence, scoring the entire read probabilistically against the reference set, and a final deconvolution step to distinguish homologous regions.