CHAMP is now available on the CosmosID-HUB.
With our next-generation human microbiome profiler, CHAMP™, we offer high-resolution taxonomic and functional profiling that is unparalleled in accuracy, precision, and coverage to maximize insights from your study.
CHAMP™ employs over 400,000 metagenome-assembled genomes (MAGs), created from a collection of more than 30,000 microbiome samples from individuals across the world. These samples were sourced from 9 different human body sites including but not limited to: gut (stool and small intestine), vagina, skin and mouth. From this extensive human microbiome data collection, we have identified almost 7000 bacteria, archaea, and eukaryote species, including many newly discovered species. This is the foundation of our microbiome analysis services.
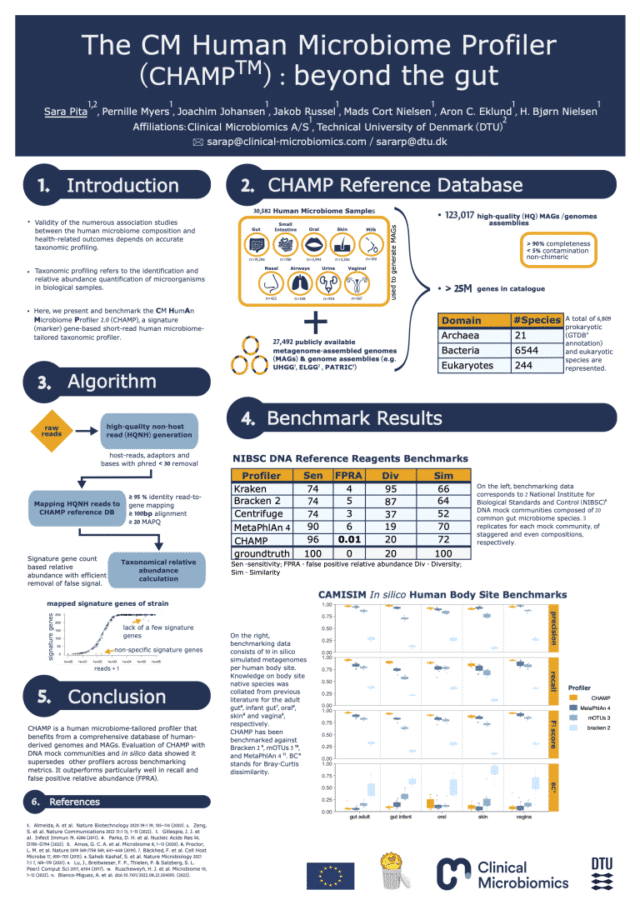
CHAMP™ provides the most accurate and sensitive microbiome profiling data thanks to the following features:
CHAMP™ demonstrates best-in-class performance using both CAMI and NIBSC benchmarks against the best and most widely used profiling pipelines in the field.
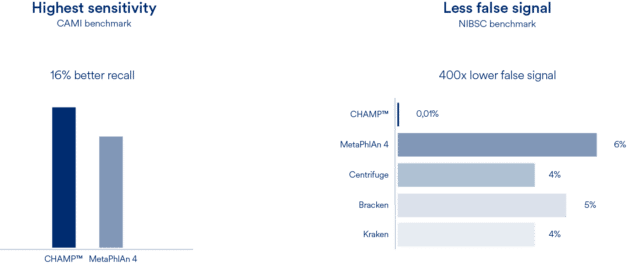
Compared to MetaPhlAn4, CHAMP™ showed 16% greater sensitivity (recall) across different human body sites and showed an astounding 400 times lower false signal in the NIBSC mock community benchmark compared to state-of-the-art profilers (MetaPhlAn4, Centrifuge, Kraken, and Bracken). This means that when CHAMP™ detects something, you can trust it’s there.
CHAMP™ uses the latest GTDB annotation which includes more rare species and, compared to NCBI, often classifies sub-species as species in their own right.
Combining the world’s most comprehensive reference catalogue for human-associated microbes with a proprietary algorithm for detection and abundance estimation of species, CHAMP™ provides you with the best microbiome data for further analysis.
For analyzing low biomass samples, searching for very rare species, profiling the microbiome of large-scale population health studies, or designing next generation probiotics or live biotherapeutics, CHAMP™ sets new standards in sensitivity and accuracy for microbiome research.
CHAMP™ also provides phage and virome profiling and advanced functional annotations. Furthermore, with the addition of our clonal-level analysis tool, it can separate very closely related strains, helping researchers understand the dynamics between probiotics/therapeutic microbes and endogenous populations of the same species.
CHAMP™ is available as part of our end-to-end human microbiome research services or as a standalone service for profiling or re-profiling of shotgun sequencing data.
Download our CHAMP™ poster presented at the EMBO | EMBL Human Microbiome Symposium 2023 in Heidelberg or contact us for more information.