


Strain tracking and engraftment persistence analysis are valuable tools to evaluate the efficacy, safety, dynamics and long-term effects of live biotherapeutic products (LBPs), fecal microbiota transplants (FMTs), and probiotics:
Strain tracking and engraftment analysis thus serve as indispensable tools, facilitating precision medicine by enabling the customization of microbial interventions based on individual responses and ensuring the optimal therapeutic outcomes for these innovative approaches in the field of microbiome-based therapeutics.
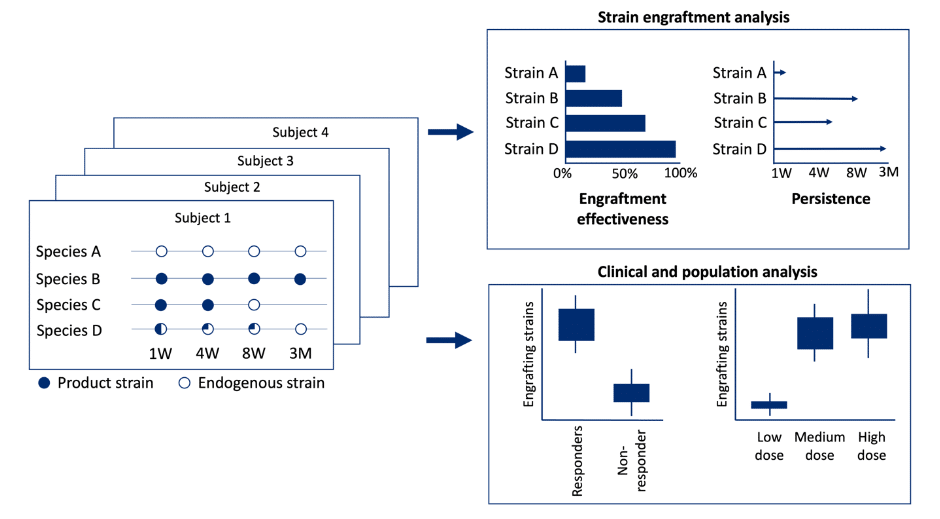
From discovery through clinical validation, strain tracking and engraftment persistence analysis help inform and understand:
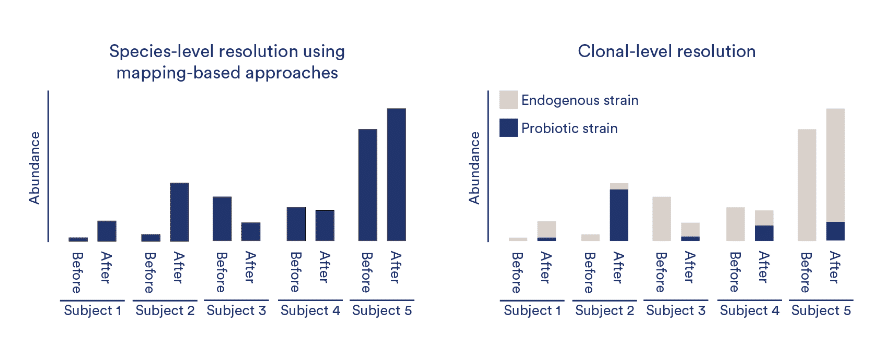
Few microbiome analysis methods can achieve the high resolution necessary for strain tracking and engraftment persistence analysis. As seen in the figure below, resolving taxonomy at the species or subspecies level will not allow you to accurately assess engraftment since it is not able to distinguish between therapeutic strains and endogenous host strains.
Clinical Microbiomics/CosmosID have developed a robust technique for de novo clonal-level microbiome profiling. This service provides you with information about clonal populations (strains) found in your samples based on variations at the single nucleotide (SNV) level.
Read our case study, based on a publication demonstrating the use our clonal-level microbiome profiling pipeline.¹
Contact us via the form to learn more!
¹ Wei, S., Jespersen, M.L., Baunwall, S.M.D. et al. Cross-generational bacterial strain transfer to an infant after fecal microbiota transplantation to a pregnant patient: a case report. Microbiome 10, 193 (2022). https://doi.org/10.1186/s40168-022-01394-w