Special thanks to Eray Şahin for the bioinformatics analysis of data.
This guide will discuss the impact of delivery mode and feeding method on infant gut microbiome establishment, a study conducted by researchers from the University of California Davis.
The research team used The CosmosID Metagenomics App to analyze microbial samples collected from infants born vaginally or through cesarean section who were also either exclusively breastfed or formula-fed.
By comparing these four groups and analyzing their associated microbiome, the team was able to gain insight into how different modes of birth and types of nutrition affect an infant’s early microbiome development.
This guide will explore these findings in detail and provide a comprehensive overview of how CosmosID helped support this important research effort.
Background
Human colonization by microbes starts as soon as the moment of birth. Events during early life can influence what microbes infants are exposed to and which of those are established in the developing gut.
Delivery mode (caesarean section or vaginal) and feeding method (breast or formula) could vary the trajectory of microbial colonization of the human gut. In fact, variation in delivery mode and feeding has been shown to perturb the gut microbiome and influence the development of asthma and atopy.
Although there is a vast amount of studies investigating the development of the human gut microbiome during the first year of life, there is a gap in knowledge regarding the development of the human gut microbiome during the neonatal period, the first month of life.
Consequently, the specific impact of different feeding methods and delivery modes on the development of the neonatal gut microbiome remains an active area of research.
Research methods
To investigate the question of the impact of delivery mode and feeding method on microbiome variation, we imported 1679 metagenomics samples from the study of Shao et al. (2019) using the accession code PRJNA313047. These fecal samples were taken from neonates delivered at 3 different UK hospitals.
We imported the whole genome sequencing data and ran those samples through the CosmosID-HUB Microbiome to illustrate strain-level resolution variation between the fecal microbiomes of breastfed (BF) or non-breastfed (NBF) neonates delivered vaginally and by C-section.
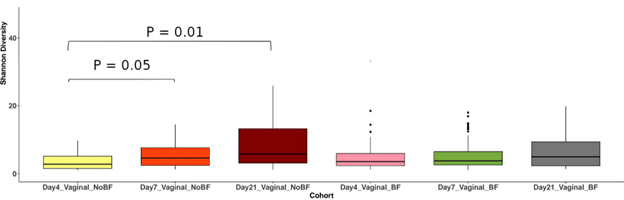
The first question to be addressed was if breastfeeding and no-breastfeeding had an impact on gut microbiome colonization of C-sections and vaginally delivered infants. We calculated the Shannon alpha diversity index of strain level resolution of breastfed and non-breastfed vaginally delivered infants at the 4th, 7th and 21st days of life (Fig. 1).
Figure 1: Non-breastfed, vaginally-delivered infants show a significant increase in alpha diversity in the first 21 days of life, while breastfed infants do not.
The box plots illustrate no significant alpha diversity difference in breastfed vaginally delivered infants during the first 21st days of life. On the other hand, we observed significant increases in Shannon’s Index of non-breastfed vaginally delivered neonate gut microbiome at the 7th (p < 0.05) and 21st (p < 0.01) days of life when compared to the 4th day.
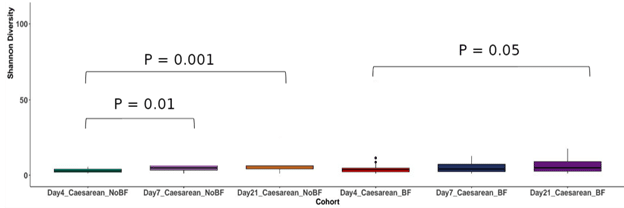
To sum up, non-breastfeeding increases species richness and evenness in vaginally delivered non-breasted infants in the 1st three weeks of life. In C-section-delivered, NBF neonates, Shannon’s Index significantly increases (p < 0.01) from the 4th day to the 7th day, but then sharply decreases on the 21st day (p < 0.001, Fig. 2).
Figure 2: Alpha diversity increases significantly from day 4 after birth in NBF and BF C-section-delivered infants.
In contrast to the vaginally delivered BF neonates, Shannon’s Diversity Index of the gut microbiomes of C-section BF infants significantly increases from day 4 to day 21 (p < 0.05).
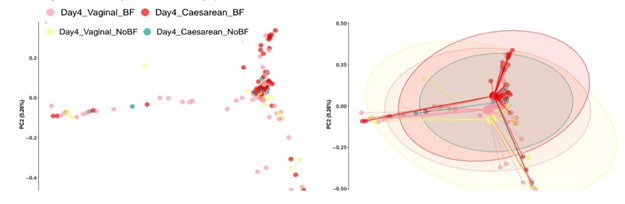
We next tested whether BF and NBF C-section and vaginally-delivered neonates harboured significantly dissimilar community compositions in microbial strains.
To do that, we compared the gut microbiome beta diversities of the infants on the 4th day of life.
Amongst the pairwise comparisons of Bray-Curtis dissimilarity matrices of vaginally delivered BF and NBF infants, as well as C-section BF and NBF infants, only the pairwise comparison between the vaginally delivered infants that are BF or NBF did not illustrate any significant dissimilarity at all (Fig. 3).
Figure 3: Only vaginally-delivered BF and NBF infants do not show significant differences in beta diversity by pairwise PERMANOVA.
This implies that breastfeeding may have a greater effect on the gut microbiome community compositions of C-section infants on the 4th day of life.
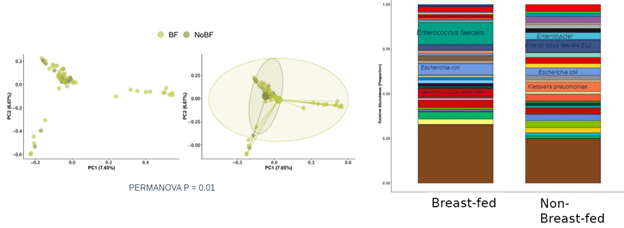
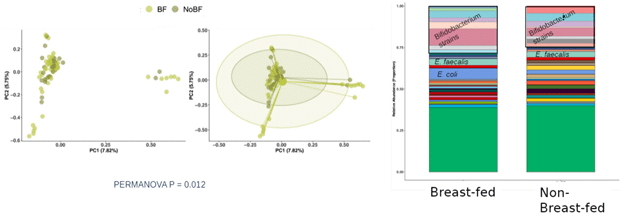
Given the significant impact of breastfeeding on C-section neonates, which are at higher risk of negative health conditions, we decided to dissect the gut microbiome variation of the C-section cohort. To do that, we calculated and plotted the beta diversities of BF and NBF C-section neonates at day 4 of life, to highlight their significant divergence.
Then, we plotted the bacterial strain relative abundances stacked bar plots to point out taxonomic differences between the two cohorts (Fig. 4)
Figure 4: Beta diversity of BF and NBF C-section delivered infants is significantly different, with BF infants showing a greater abundance of Enterococcus faecalis and E. coli but greater potential pathogen carriage in NBF infants.
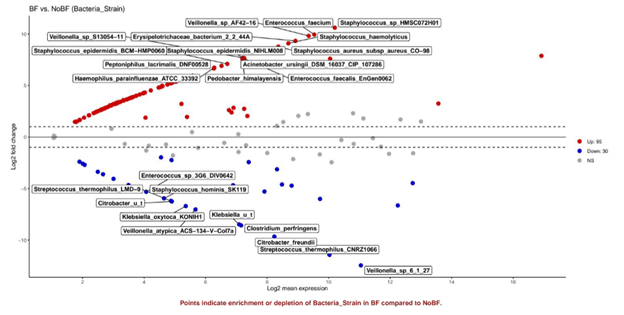
The stacked bar plot illustrates that, while breastfed C-section infants contain strains of Enterococcus faecalis and E. coli, non-breast-fed C-section infants appear to carry more Gram-negative strains. To determine if these observed differences are significant, we performed different abundance analyses using DESeq2 (Fig. 5)
Figure 5: More taxa are enriched in BF (red) infants than in BF infants (blue).
The volcano plot illustrated that more species were differentially abundant in breastfed infants (95 > 30) and confirmed the observation from the relative abundance box plot on the higher abundance of E. faecalis in BF infants.
The next question we examined was whether these differences persisted to the 21st day of life. To do that, we again compared relative abundances using box plots. Comparing the relative abundances illustrated that E. coli is lower in NBF infants, as also observed on the 4th day, and Bifidobacterium strains are present in both BF and NBF infants.
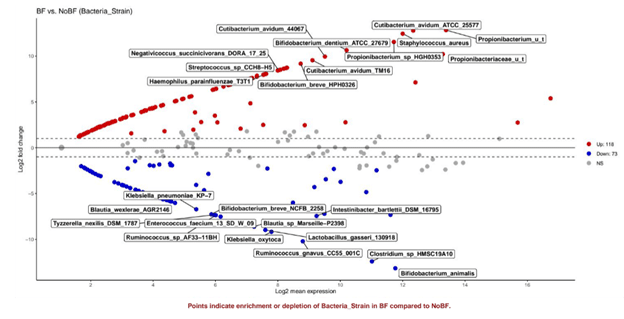
However, beta diversity is not significantly different between BF and NBF infants at 21 days after birth (Fig. 6). To determine whether specific taxa were differentially abundant between the groups, we again performed a differential abundance analysis using DESeq2. The volcano plot (Fig. 7) illustrated that, as of day 4, BF infants had more differentially abundant taxa than NBF infants (118 > 73).
Figure 6: Beta diversity is not significantly different at day 21 between C-section BF and NBF infants; Bifidobacterium strains can be detected in both groups.
Figure 7: More taxa are significantly enriched in BF (red) than in NBF (blue) C-section infants; Bifidobacterium strains are significantly enriched in both groups.
Unlike at day 4, several Bifidobacterium strains are significantly enriched in both the BF and NBF infants at day 21, Bifidobacterium breve HP0326 and Bifidobacterium dentium ATCC 27679 are enriched in BF infants and Bifidobacterium animalis and Bifidobacterium breve NCFB2253 were differentially abundant in NBF infants.
This suggests that important Bifidobacterium species are able to colonize the C-section-delivered infant within the first month of life, regardless of feeding method.
Outcomes
In conclusion, the comparative analysis of BF and NBF infants’ gut microbiomes through the first month of life illustrates key differences.
We focused on infants delivered by C-section, who tend to be at higher risk for adverse health effects, and saw that the gut microbiome of these babies is significantly influenced by feeding methods within the first four days of life.
However, by day 21, these differences become non-significant and strains of Bifidobacterium breve colonize both BF and NBF infants.
This suggests that, while there may be early differences in these infants based on feeding method, ultimately their gut microbiota are colonized by Bifidobacterium strains important in the maturation and development of the infant microbiome. Longitudinal sampling is necessary to understand whether these early differences are associated with life-long health impacts.
Wider impact
As evidenced in this work, the early-life gut microbiome is an important factor for health outcomes later in life. It is therefore essential to understand what influences the infant microbiome and how it may be altered or even corrected if needed.
This study sheds light on the differences between BF and NBF infants delivered by C-section, showing that while there may be early differences in the gut microbiota, ultimately Bifidobacterium strains colonize both BF and NBF infants.
This has implications for public health initiatives which aim to reduce adverse health outcomes by optimizing infant feeding methods and microbiome development. It also highlights the importance of longitudinal sampling as a means to understand any potential long-term health impacts associated with early-life microbiome differences.
Ultimately, this work provides a basis for further research into the mechanisms behind infant microbiome development and its implications for health outcomes later in life.
How did CosmosID support this research?
The CosmosID-HUB microbiome analysis platform offered a comprehensive and powerful tool that was instrumental to this research. It afforded the team the ability to analyze large amounts of data quickly and accurately, enabling them to compare the gut microbiomes of BF and NBF infants delivered by C-section.
The CosmosID platform and state-of-the-art microbiome sequencing services allowed the researchers to identify differentially abundant taxa between BF and NBF infants, as well as to identify correlation patterns. Additionally, the platform enabled the team to explore associations between taxa at different time points and infer early-life microbiome development trajectories.
The researchers also took advantage of CosmosID’s interactive visualization tools to understand their data better. By creating interactive graphs, they were able to compare beta diversity between BF and NBF infants delivered by C-section, as well as to identify differentially abundant taxa.
Unlock the power of the microbiome with CosmosID. Get started today.
Want more like this? Sign-up to our newsletter to get the latest news from CosmosID: