Background
Metagenomics studies using shotgun sequencing have been an important tool for investigating the gut microbiome of pets, including dogs, in recent years.
These studies involve sequencing all DNA fragments present in a sample, allowing for the identification and characterization of the diverse range of microorganisms present in the gut microbiome. Such studies have revealed the dog gut microbiome’s complexity and provided insights into the impact of diet on its composition (Beloshapka et al., 2013).
Furthermore, 16S rRNA sequencing of fecal material from normal-weight and obese dogs illustrated significant differences in bacterial taxa at a phylum level—moreover, significantly different functional gene compositions between the two dog cohorts were predicted (Thompson et al., 2022).
Clearly, the gut microbiome has implications for dog health, and care. By utilizing metagenomics sequencing, researchers can gain a better understanding of the dog gut microbiome and the impact of dietary interventions, ultimately leading to the development of targeted nutritional strategies for improving the health and well-being of dogs as pets.
Another recent study that reflects on dog care illustrates that a pet’s favorite soft, wet food may not be as good for your beagle as it thinks it is.
A recent study compared the cortisol levels and gut microbiomes of beagles that were fed dry and water-softened food. The results showed elevated cortisol levels and enrichment of opportunistic pathogens in the gut microbiome of beagles fed wet food, along with an increase in gut microbiome alpha diversity. (Zhang et al., 2022).
This increase in opportunistic pathogens, in combination with the cortisol increase, suggests that wet food may not be the healthiest choice for your dog.
Latest research
To expand on the impact of dietary variation on the beagle microbiome, we conducted an analysis of a study in which beagles were fed either a high-protein, low-carbohydrate food or a low-protein, high-carbohydrate food. Although the study included labradors and beagles, we limited our analysis to the beagles to minimize the impact of host genetic variation.
The original study collected feces from 16 beagles in each diet group, for a total of 32 dogs. DNA was extracted and sequenced using whole genome shotgun sequencing. We accessed the publicly available sequencing data using the accession code PRJEB20308 (Coelho et al., 2019).
Then, we imported the data using the Sequence Read Archive importer tool on the CosmosID-HUB, filtered the data, and analyzed the microbiome variation.
Results
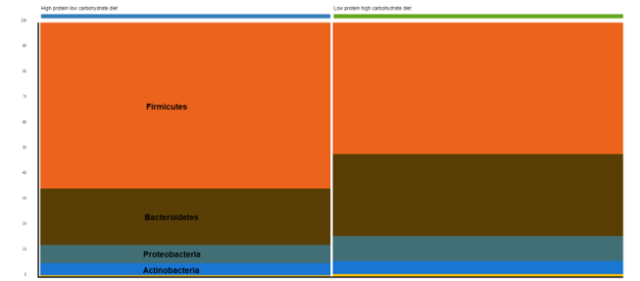
Bacterial phyla relative abundance, shown in the stacked bar plots in Figure 1, illustrates the enrichment of Bacteroidetes and Proteobacteria with the low-protein, high-carbohydrate gut microbiome, while strictly anaerobic Firmicutes were enriched in the gut microbiome of high-protein, low-carbohydrate diet-fed beagles.
Figure 1: Phylum-level relative abundance of the gut microbiome in high-protein, low-carb-fed dogs and low-protein, high-carb-fed dogs.
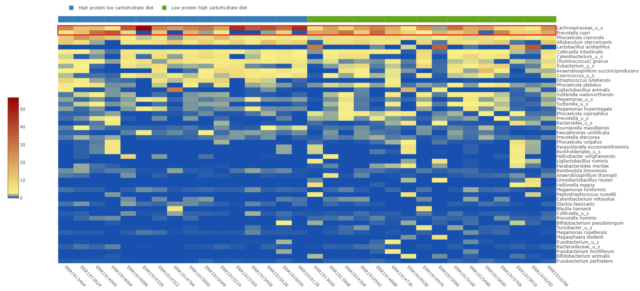
At the species level, illustrated in the heatmap in Figure 2, an unidentified species in the Lachnospiraceae family seems to be driving the enrichment of Firmicutes in the high-protein, low-carbohydrate diet-fed beagles. An increase in Prevotella copri relative abundance in the low-protein, high-carbohydrate diet-fed beagles microbiome appears to be responsible for the elevated Bacteroidetes phyla within this group.
Expansion of Prevotella copri relative abundance is known to correlate with adverse phenotypes such as arthritis susceptibility (Scher et al., 2013) as well as non-alcoholic steatohepatitis development in obese subjects (Moran-Ramos et al., 2023). It is possible that this diet-induced increase in beagles may be associated with poor health outcomes as well.
Figure 2: Species-level relative abundance of the gut microbiome per dog sorted by diet.
To investigate significant differences in species richness and evenness, we compared the Simpson’s, Shannon’s and CHAO1 alpha diversity indices between the diet cohorts. Although all three indices emerged to be higher in low-protein, high-carbohydrate diet-fed beagles, only the CHAO1 index illustrated significant results using the Wilcoxon Rank Sum Test (p < 0.04, Figure 3). These results suggest that species evenness is similar between the diet groups but that they differ significantly by richness or the number of different species present. This could be due to the selection of specific taxa by the dietary components.
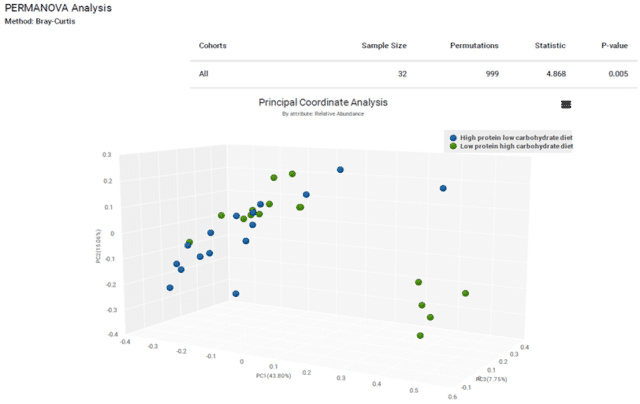
A comparison of the Bray-Curtis diversity indices of the two groups showed significant differences in community composition (p < 0.005, Figure 4). While alpha diversity shows significant differences by number of species present, beta diversity shows significant differences in overall community structure. This provides further evidence that the different diets select for specific bacterial taxa, such as Prevotella copri and Lachnospiraceae. Longitudinal sampling of these dogs on these diets could reveal specific health impacts linked to these microbiome shifts.
Figure 4: Bray-Curtis beta diversity is significantly different by diet.
Conclusions
In summary, our analysis illustrated that the fecal microbiome of low-protein, high-carbohydrate-fed beagles had fewer strict anaerobes. This suggests an anaerobic shift in the gut, which may have enriched for Prevotella copri, which is associated with multiple adverse phenotypes. Further, we saw a significant increase in species richness in this diet group, which is associated with increases in opportunistic pathogen carriage and stress in beagles.
From this point of view, the low-protein, high-carbohydrate diet seems to have fewer potential health benefits for beagles.
However, future studies should be conducted to assess the long-term impact of this diet on beagle health outcomes. These studies should include assessment of host response, such as through immune profiling and metabolomics, in order to create a better understanding of the microbiome-host interaction in response to dietary changes in the beagle.
In conclusion, our results suggest it may be good for your beagle to avoid carbohydrate-rich diets, but please consult your veterinarian if you have questions on how to feed your best friend.
How did CosmosID support this research?
The CosmosID-Hub was used in this study to provide a comprehensive platform for data analysis, visualization, and bioinformatics tools. Using the Hub to analyze and visualize the microbiome composition of our beagle cohorts by different taxonomic levels, this study was able to uncover significant differences between the two dietary cohorts.
At CosmosID, we’re proud to support research projects in all sectors with our microbiome sequencing services and bioinformatics services. Our state-of-the-art tools and analysis capabilities provide researchers with the data necessary to make informed decisions.
We’re inspired by the results of this research and looking forward to continued collaborations in microbiome research projects like this in the future!
Looking for results like these? Get started today.
Want more like this? Sign-up to our newsletter to get the latest news from CosmosID: