By Barış Özdinç
Dogs have been man’s best friend for centuries — and continue to be popular pets today. According to the American Pet Products Association National Pet Owners Survey, 53.4% of American households own a dog.
People treat their dogs like family members, including looking after and investigating ways in which to improve their furry companion’s health and longevity. When it comes to the health of our beloved canine companions, the Labrador and the Beagle, one aspect of their health includes the state of their gut microbiome.
The gut microbiome has known impacts on human health, including body weight, immunity, and metabolism. It has similar impacts on dog health (Pilla 2019) and may even influence the dog’s owner’s microbiome, especially if that owner is an infant (Tun 2017).
A robust and diverse gut microbiome plays a pivotal role in their overall well-being, fostering a robust immune system while keeping pathogenic bacteria at bay. This article delves into the fascinating world of dog microbiome, particularly focusing on factors that affect the gut health of Labradors and Beagles.
Understanding the Significance of Gut Bacteria and Microbes in Canines
Gut bacteria, along with other gut microbes, facilitate vital functions such as nutrient absorption, immunity, and even mental well-being.
A harmonious balance of these bacteria is crucial to maintain a healthy state in dogs. However, any imbalance, often reflected in the form of an overgrowth of pathogenic bacteria, could possibly lead to conditions such as idiopathic inflammatory bowel disease (IIBD).
In dogs, IIBD manifests in the form of chronic intestinal inflammation, causing symptoms such as diarrhea, weight loss, and in severe cases, even death. Thus, it is essential to pay attention to gut health in dogs and take preventive measures to maintain a healthy gut microbiome.
Short Chain Fatty Acids – The Unsung Heroes
One prominent factor contributing to gut health is the presence of short chain fatty acids (SCFAs).
These are produced when gut bacteria ferment dietary fibers in the canine intestinal tract. SCFAs play a crucial role in maintaining gut health, acting as a primary energy source for colon cells and promoting a healthy intestinal environment that resists pathogenic invasions.
The Gut Health Dichotomy: Labradors vs Beagles
While Labradors and Beagles belong to the same canine family, their gut microbiome can vary. This variation may be due to factors such as diet, genetics, and environment. For instance, dogs fed a certain diet may have a different fecal microbiome compared to those on another diet.
Labradors and Gut Health
Interestingly, Labradors are often prone to conditions such as obesity and IIBD. Their predisposition towards these ailments has often been linked to their gut microbiome. A diet rich in fiber, promoting the production of SCFAs, could potentially support a healthier gut microbiome in Labradors.
Beagles and Gut Health
On the other hand, Beagles are typically known for their robust health. However, they are not immune to gut health issues. IIBD, although less common than in Labradors, can still surface in Beagles due to an imbalance in their gut bacteria.
Fecal Microbiota Transplantation – A Ray of Hope
In both Labradors and Beagles, fecal microbiota transplantation (FMT) has shown promising results in treating IIBD. The procedure involves introducing a healthy fecal microbiome from a donor into the gut of a sick dog to restore the balance of gut bacteria.
Canine Gut Microbiome Composition: Our Study
It is known that human genetics can influence the gut microbiome, and a previous study indicates that different dog breeds, which differ genetically, may have specific gut microbiota signatures (Reddy 2019). In our microbiome dataset comparison this week, we look at gut microbiome differences between 32 beagles and 32 Labradors.
This data comes from a study by Coelho et al. (2018), who fed the dogs the same baseline diet. We imported the publicly available data using the Sequence Read Archive importer tool on the CosmosID-HUB platform, filtered the data, and analyzed the microbiome variation.
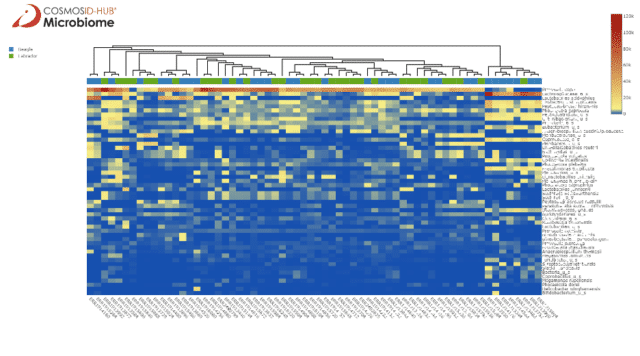
Comparing the two breeds bacterial species abundance scores in the heatmap (Figure 1) illustrates an intermixing of the samples, highlighting that dog breed may not be strongly associated with gut bacteriome variation, at least at the species level.
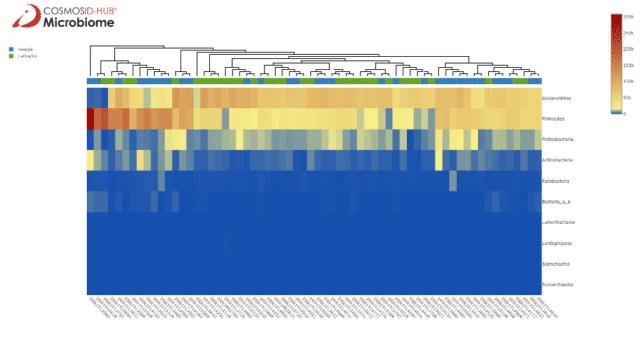
There is an interesting cluster on the right end of the heatmap composed of mainly beagle samples with high abundance of Lachnospiraceae. When we instead look at the phylum level in Figure 2 we see some clustering by breed, with labradors having more Bacteroidetes and beagles having more Firmicutes. On either end of the heatmap are clusters with mostly beagle samples, one with similar Bacteroidetes and Firmicutes abundances and another with extreme Firmicutes abundance but lower Bacteroidetes.
Although the dogs started the study on the same diet, they differed in sex, age, and body weight. One of these confounding factors may influence the clustering we observe among the beagle microbiome.
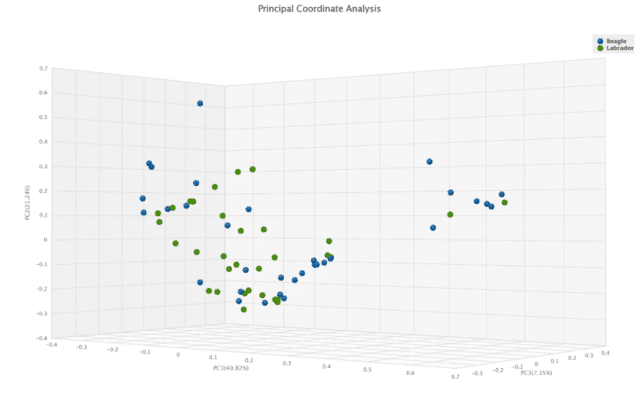
We next performed alpha and beta diversity testing on the samples to identify significant differences in diversity between the breeds. None of the tested alpha diversity indexes (CHAO1, Simpson and Shannon) were significantly different, highlighting that beagles and labradors have similar amounts of different species with similar abundance in their guts.
Investigating beta diversity differences provides a slightly different story. Here, the Jaccard dissimilarity matrix comparison of the two breeds showed a difference (p = 0.021), implying that some taxa are present or absent only in one breed or the other. The Bray-Curtis dissimilarity matrix was not significant (p = 0.059, Figure 3).
This suggests that the significance found by the Jaccard index may be driven by low-abundance species that may not drive an overall difference between the communities.
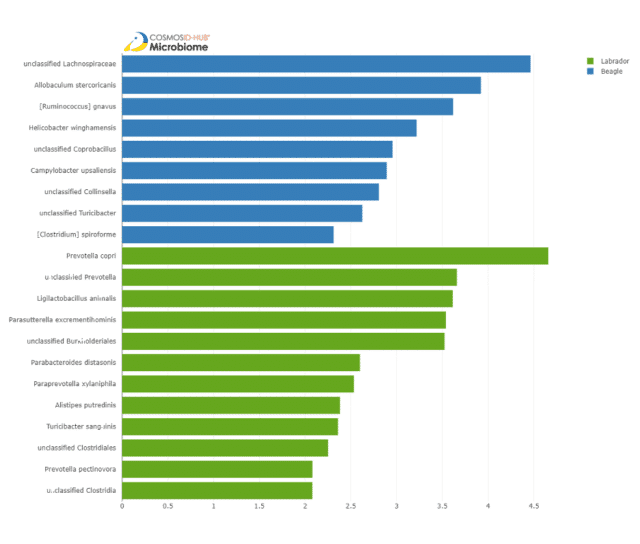
To identify specific taxa that may be significantly different between the breeds, we used the differential abundance tool LEfSe (Linear discriminant analysis effect size). LefSe identified 9 differentially abundant species in beagles and 13 in labradors (Figure 4).
These results suggest that beagles may be prone to gastroenteritis due to significant enrichment of Helicobacter winghamensis and Campylobacter upsaliensis. These bacteria are known agents of gastroenteritis in dogs, as well as gastrointestinal disorders in humans (Solnick et al. 2002, Bourke et al. 1998).
Species that are differentially abundant in labradors include:
- Prevotella copri and Prevotella pectiovora: which are associated with plant-rich diets (Kovatcheva-Datchary et al. 2015);
- Parabacteroides distasonis: which may alleviate obesity and metabolic dysfunction (Wang et al. 2019);
- Paraprevotella xylaniphila: which is associated with an active lifestyle (Bressa et al. 2017); and
- Turicibacter sanguinis: which plays important roles in serotonin metabolism in humans (Fung et al. 2019).
Our study: Conclusion
To conclude, LefSe analysis suggests that beagles have more enrichment of dysbiotic species, but labradors have more beneficial bacteria enriched in their guts. These findings show that the gut microbiome may differ by dog breed and influence the health of the animal. In this comparison we have not taken into account any differences by dog age, sex, or body weight.
These factors are known to influence the gut microbiome in humans and are likely to do so in dogs as well. In a later update we will explore dog microbiome variation by these factors and determine their influence on the differences we found this week. Stay tuned for more!
As for now, we hope you enjoyed the insights from our study. We believe that understanding the gut microbiome of dogs can help improve their health and well-being, as well as deepen our knowledge of the human gut microbiome. Thank you for reading! Happy pet parenting! 🐾
Unlock the Power of the Microbiome with the CosmosID-HUB
At CosmosID, we are dedicated to unlocking the power of the microbiome through cutting-edge technology and groundbreaking research. The CosmosID-HUB platform provides a comprehensive analysis of microbial communities in various environments, including humans and animals.
By utilizing advanced bioinformatics and machine learning algorithms, our team is able to accurately identify and quantify microorganisms at the strain level, providing valuable insights into the diversity and function of the microbiome.
Our platform is widely used in research, clinical, and environmental settings to understand the role of the microbiome in health and disease.
If you are interested in learning more about our HUB platform or partnering with us for your next microbiome study, reach out to our consultantstoday.


