In the captivating world of microbiology, two primary methods have emerged to study the composition, structure, and function of microbial communities: 16S ribosomal RNA (rRNA) gene sequencing and metagenomics.
This article will delve into a comparative analysis of these two techniques. We will explore their fundamental principles and methodologies of each sequencing method, before looking at the key similarities, differences, and the unique insights they offer into the microbial world.
The goal is to empower you, the reader, with a comprehensive understanding of the differences between 16S and metagenomics, thus enabling you to make an informed choice between these two potent tools.
What is 16s rRNA gene sequencing?
16S rRNA Amplicon sequencing is the selection and isolation of the 16S gene region of interest from before sequencing to illustrate the genetic variation between the samples within the target region.
What this essentially means is that targeted sequencing of the 16S gene region captures the genetic variation of microbial communities in DNA sequences — specifically within the known 16S genetic region which can then be compared between samples.
What is shotgun metagenomic sequencing?
In shotgun metagenomics, whole DNA is extracted from a sample then exposed to random fragmentation prior to NGS. Thus, enabling strain-level multi-kingdom taxonomic classification, functional profile characterization, and AMR detection.
By taking an untargeted approach to sequencing whole DNA and avoiding primer-dependent PCR Amplification used in amplicon sequencing services such as 16S, one gains an exponential amount of data and advantages alongside it.
As microbial diversity is vast and complex, the importance of metagenomics as an approach for studying it becomes clear.
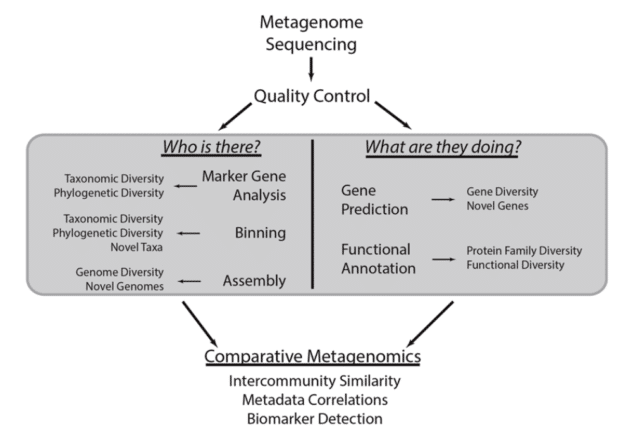
Frontiers – FIGURE 1. COMMON METAGENOMIC ANALYTICAL STRATEGIES. This methodological workflow illustrates a typical metagenomic analysis.
Shallow shotgun sequencing
In shallow shotgun sequencing, the shotgun metagenomic approach is applied to DNA from samples, however, the sequencing depth targets of these samples are much lower than standard metagenomic sequencing. In doing so, the amount of data generated for each sample is reduced while also reducing the sequencing costs per sample.
The DNA extraction method of choice often depends on the type of sample being studied. For microbial samples, DNA can be extracted using the standard methods such as commercial collection kits.
While the data generated from shallow shotgun sequencing is not as robust as deep shotgun sequencing, the data generated still enables a multi-kingdom species/strain resolution approach to various sample types, thus offering a cost-optimized solution for researchers wanting to have greater insights into the microbial community when compared to 16S sequencing.
You Might Like: Battle of the Sequencers: Nanopore vs Illumina Sequencing Technologies
What’s the difference between 16s and shotgun metagenomics?
While both 16S rRNA sequencing and shotgun metagenomic sequencing have the ability to characterize the microbiome via next-generation sequencing (NGS), there are also a great deal of differences between the two that are worth noting.
16S rRNA sequencing primarily utilizes PCR to amplify the specific gene region of interest (V4, V9, etc) that is then templated for NGS, thus only 16S gene sequences are generated. This is contrary to shotgun metagenomics, in which the entirety of genomic content is templated for NGS, thus a much more broad representation of microbial genomes are present for characterization.
As a result of this core difference in molecular processes, there are a great number of differences between these approaches that are worth noting when considering which approach is applicable for your research. These differences are detailed below:
Taxonomy resolution
- 16S rRNA: Family & Genus level of bacteria, Species level possible however there is high rate of false positives
- Shotgun Metagenomics: Species and Strain level resolution of multi-kingdom (bacteria, virus, fungi, protist)
Functional profiling
- 16S: Directly, 16S sequencing provides only taxonomic information. Functional profiles can be inferred based on known functions of less abundant taxa, but this is an indirect approach and may not capture the true functional diversity.
- Shotgun Metagenomics: Directly provides information on the functional genes and pathways present in the microbial community. It can capture both known and novel microbial marker genes, offering a more comprehensive view of the functional potential of the community.
Recommended sample type
- 16S rRNA: All sample types are applicable for this technique. Areas in which this technique has increased utility are in samples with low microbial biomass and/or high non-microbial biomass present within the sample. This includes samples such as skin swabs, environmental swabs, etc.
- Shotgun Metagenomics: All sample types are applicable for this technique. In samples such as stool, shallow shotgun sequencing is much more appropriate in that this sample type has a high microbial biomass with low non-microbial content.
Multi-Kingdom coverage
- 16S rRNA: Only covers bacterial characterization. Additionally, depending on the bacteria in which you would like to differentiate, there will be a more appropriate variable region of the 16S gene to target (V4, V9, V1V3, etc).
- Shotgun Metagenomics: This approach allows for multi-kingdom characterization without any adjustments/customization. Bacteria, Fungi, Virus, and Protist are characterized using this technique.
Host DNA interference
- 16S rRNA: Host DNA interference does not occur, as this approach utilizes PCR to amplify and template the gene region of interest (16S rRNA), thus removing host DNA presence and only generating sequencing data of the 16S gene.
- Shotgun Metagenomics: Host DNA can impact the quality of analysis from shotgun sequencing data, as an increase in Host DNA will decrease the signal of microbial DNA signatures within the data. Thus, increased sequencing depth OR Host DNA removal techniques are recommended when dealing with samples of high Host DNA.
Cost per sample
- 16S rRNA: Costs are lower per sample when compared to shotgun metagenomics, as the amount of sequencing data required to generate a serviceable dataset is much lower than microbial genomic databases.
- Shotgun Metagenomics: If the sample type is appropriate for shallow shotgun sequencing, the costs can be quite close to 16S rRNA sequencing. However, if the sample type is more complex, or greater investigational interests are involved with the research, then deeper sequencing (and thus additional cost) will be required.
Minimum DNA input
- 16S rRNA: As this approach is PCR based, the DNA input required for this reaction is minimal, with successful reactions from less than 1 ng of DNA being a routine occurrence.
- Shotgun Metagenomics: As this approach utilizes the sequencing of all genomic DNA content, the DNA input for this reaction is much greater than 16S rRNA sequencing. Typically, samples are requested to be at minimum 1ng/uL for shotgun metagenomics, however additional techniques for low biomass samples are available to be utilized if necessary.
16S sequencing vs shotgun metagenomic sequencing: which is right for you?
With this information in mind, it is clear that both 16S rRNA sequencing and shotgun metagenomic sequencing have their applications and advantages. Thus, when considering your research needs, keep these factors in mind:
- Does my sample have low microbial content/high host content?
- Do I need a generic characterization of the bacterial community, or do I need species/multi-kingdom/functional pathway characterization?
Consult an expert today and unlock the power of the microbiome.
At CosmosID, we have experts with years of experience in microbiome methods ready to consult you on your research needs.
Our leading shotgun metagenomics & custom bioinformatics pipelines are powered by the latest technology and provide you with unmatched qualitative and quantitative insights into your data, unlocking new possibilities in your microbiome research.
Learn more about our 16S Amplicon as well as Shallow and Deep Shotgun Metagenomics solutions today, and if you have any questions don’t hesitate to contact us today.
16S vs metagenomics FAQs
Is 16S or shotgun sequencing best for the microbiome?
Unfortunately, the answer here is it depends! You need to assess the qualities of the specific sample type that you are working with, as well as the level of resolution and insight your research needs when it comes to the microbiome analysis.
If you need assistance on figuring out which application is the best for you to understand and identify bacteria or DNA, don’t hesitate to contact one of our microbiome experts and we’d be happy to assist!
Why do we choose 16S rRNA instead of others to identify microorganisms?
The 16S rRNA gene has been identified as a highly conserved gene within the Bacterial kingdom with taxonomically relevant mutations between bacterial organisms. Thus, by amplifying this specific gene region, the mutations in this gene allows for the differentiation of various bacterial organisms within a sample.
Want more like this? Sign-up to our newsletter to get the latest news from CosmosID: