Throughout the years, microbiology has continuously pushed the limits to test for, detect, and measure the microbes found in various environments. However, as the questions surrounding microbiology, and the role microbes play in various aspects of life, the methods in which microbes are tested for and measured needed to improve.
One of these questions that microbiology is dealing with today is that of the microbiome, which can be described as the total community of microorganisms within an environment.
Today’s researchers not only want to understand all of the organisms that are present within the microbiome, they want to understand the community proportions that the various organisms occupy, the functional capacity of the microorganisms within the microbiome, and so much more.
Thus, in order to address all of these questions, microbiology has moved towards a powerful tool that can provide answers: Next-Generation Sequencing.
Next-Generation Sequencing (NGS) is an important advancement in the field of microbiology, as this approach enables the total genetic content within a sample to be transcribed into data that can be further analyzed for microbiome characterization (microbial ID, abundance measurements, gene detection, etc.).
By using this technique, all microorganisms within a sample can be detected and measured for researchers, thus providing a full picture of the microbial environment to be studied. This is also important for microbiome research, as many microorganisms within a sample may have various growth conditions, etc. that could prove difficult to handle with traditional microbiology techniques.
By using NGS, all organisms are subjected to DNA isolation and sequencing, thus improving sensitivity and coverage to all microorganisms within a sample.
In this article, our microbiome scientists will take a deeper look at how to sequence a microbiome, all of the steps involved in microbiome sequencing, and the challenges (and relative solutions) that microbial sequencing brings about.
What is Microbiome Sequencing?
As mentioned above, the microbiome is the total community of microorganisms within a given environment. With the microbiome consisting of such a diverse set of microorganisms, traditional microbiology methods such as culture lack the efficiency, coverage, and power necessary to contextualize the entirety of the microbial population found within samples.
Thus, as NGS has become increasingly more popular, less expensive, and more applicable to microbiome research, sequencing has become the primary solution to studying the microbiome. By utilizing microbiome sequencing, questions in microbiology can be answered faster, with more information, and with more coverage of microbes within samples than ever before.
Whether this is trying to understand microbe-microbe interactions between bacteria, fungi, or viruses, or if it is to capture resistance genes present within microbial communities, or the overall functional pathways within a sample, microbiome sequencing is a powerful tool that enables various applications.
These applications include screening the human microbiome (gut, oral, skin, vaginal, etc) for research or clinical studies, testing wastewater for harmful pathogens and epidemiological monitoring, testing soil for microbial composition and gene presence in agriculture, just to name a few.
By implementing microbiome sequencing, a whole new world of answers can be found to characterize the microbes present within your samples, thus enabling your research to be much more comprehensive and insightful.
History of Microbiology & Sequencing Methods: Who Was First?
The concept of the human microbiome was first suggested by Joshua Lederberg, who coined the term “microbiome” to signify the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space” (Lederberg and McCray 2001).
However, prior to Dr. Lederberg’s coinage of “microbiome”, efforts to determine the numbers of microbes in a community and their phylogenetic relationships were well underway, which consisted of analyzing the relatively well-conserved 16S rRNA genes in mixtures of organisms, from 1977–1996.
An early method of microbiome sequencing, 16S rRNA was a milestone in the field, previously pioneered by Carl Woese, Norman Pace and others to identify environmental bacteria, based on sequencing small subunit ribosomal RNA genes (16S rRNA).
Using this approach, Wilson and Blitchington compared the diversity of cultivated and non-cultivated bacteria within a human fecal sample via sequencing methods in 1996.
Since then, the development of massively parallel sequencing, combined with whole genome shotgun sequencing, have further advanced the field of microbiome sequencing into an even more accurate and sensitive testing technique that enables the detection of all microorganisms and genes within a sample for analysis.
How to Sequence a Microbiome in 5 Steps
Step 1: Sample Collection
The first step to sequencing microbes involves the collection process. This is a very critical part in this process, as samples must be preserved to be as close to the point of collection as possible, otherwise biases and unforeseen variability of the microbiome within your samples may occur.
Thus, there are a few ways in which sample collection can be made to ensure the stability of your samples for proper microbiome sequencing, which include:
- Flash Freezing: Upon collecting your sample in a sterile container, immediately store your sample in dry ice or -80C conditions.
- Microbiome Preservation Media: Upon collecting your sample, transfer the material into a designed microbiome preservation device. This device consists of a buffer designed to preserve the nucleic acid integrity of your sample, while also preventing changes in the microbiome. CosmosID has multiple offerings for collection kits that utilize preservation media.
Step 2: DNA Extraction
Once samples have been collected, DNA extraction must be performed in a robust process to ensure that all DNA from all microorganisms within the sample is collected for downstream sequencing.
One common concern with microbiome testing is the ability to detect all gram-positive microorganisms, due to the fact that these cells are much harder to lyse open and expose the DNA.
Thus, a method of both chemical and physical lysis during DNA extraction is most optimal for sample processing, with CosmosID offering multiple extraction methods optimized to various sample types for maximum DNA yield.
Step 3: Library Preparation
Once DNA has been isolated, DNA must be properly templated and prepared for the relevant NGS technique that will be used in generating sequenced data. There are 2 main approaches that are typically used in microbiome sequencing:
- Amplicon Preparation: This technique will take the total DNA isolated from DNA extraction and amplify specific regions of interest – such as the 16S rRNA gene or ITS – these regions are amplified and barcoded for sequencing, while all other non-target genomic content is discarded.
- Whole Genome Shotgun Preparation: This DNA sequencing technique will take the total DNA isolated from DNA extraction and fragment/template all present DNA.
Step 4: Sequencing
After templating the DNA into sequence-ready material, samples are pooled together for NGS sequencing, typically using Illumina-based sequencing technologies and Sequencing-By-Synthesis (SBS) chemistries.
In this process, DNA from your samples are loaded into your chosen sequencing technology, where each DNA strand becomes denatured into single-stranded DNA fragments and complementary DNA strands are built alongside the single-stranded molecules.
In the complementary strand building process, each individual base pair that is added fluoresces a specific color, with each color signifying the particular complementary base that was added to the DNA fragment.
As these complementary bases are added to the template single-stranded DNA fragments, an optical system within the DNA sequencer captures these flashes of color, which then converts these colors into DNA sequence data for software tools to ultimately perform analysis.
Step 5: Data Analysis & Visualization
Once data has been generated through NGS, data analysis finally provides the big picture of the microbiome within the sample. There are multiple methods in which NGS data can be used to characterize microorganisms and genes within samples, with the two main methods being the following:
- Reference-Based: NGS data is mapped to known markers from a reference database.
- Metagenomic Assembly-Based: NGS data is assembled agnostically in an attempt to rebuild microbial genomes within the sample.
After the analysis/characterization of microorganisms within the sample, various analyses can be performed to investigate the microbiome relative to your research questions. These include comparative analyses, alpha/beta diversity calculations, statistical analyses, etc.
For a comprehensive microbiome analysis, CosmosID has developed the CosmosID-HUB, which provides a fully customizable, easy to use bioinformatics suite to analyze your microbiome samples.
Learn more about the CosmosID-HUB today
Challenges and Solutions in Microbiome Sequencing
Microbiome sequencing is a constantly evolving field with no signs of slowing down. While many innovations to microbiome research find a way to improve the science, it also adds to the complexity of research projects, including what you should consider when designing your own microbiome research.
Below, we have outlined a few examples of challenges in microbiome solutions, as well as proposed solutions to these challenges:
Challenge: Detection Sensitivity of Microorganisms in NGS
Solution:
- Implement sufficient DNA extraction techniques that can isolate nucleic acids of interest.
- Implement sufficient sequencing technique (Amplicon/Shotgun WGS) for detection (consider resolution needed, sample type.)
- Implement sufficient data analysis solutions with proper accuracy and specificity — like the CosmosID-HUB Award Winning Analysis Pipeline.
Challenge: Wet-Lab Efforts of Microbiome Sequencing Complex and Prone to Issues
Solution: CosmosID’s CLIA certified laboratory facility, located in Germantown, MD, provides the most robust and highest quality control system for your microbiome research to ensure the integrity of your samples during testing.
This includes running controls, maintaining properly trained staff, and rigorous review of QC metrics on a continuous basis. Additionally, CosmosID has scaled the sophisticated process of NGS testing, which allows us to offer consistent turnaround times (as fast as 3-5 days) for all your testing needs.
Challenge: Analysis of Microbiome Sequencing Data is Laborious and Complex
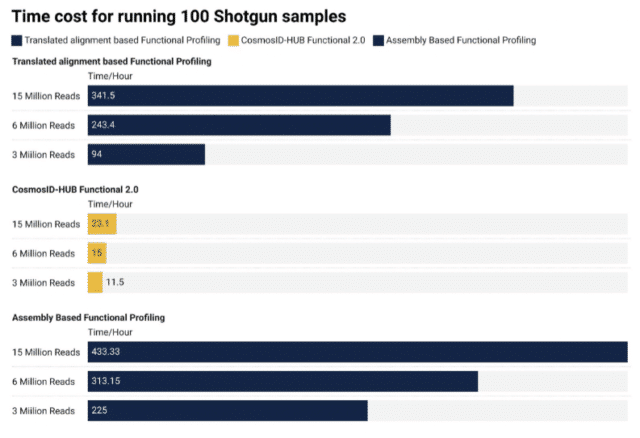
Solution: By significantly reducing the time needed to analyze data, answers to the microbiome within your sample are available much faster than any other tool available.
The CosmosID-HUB has been designed to enable analysis of microbiome sequencing data in a fraction of the time other tools take per sample, as explained in the image below:
Additionally, the CosmosID-HUB provides a dashboard suite of various analyses that you can perform on your samples, including comparative analyses, functional pathway analysis, statistical analyses, all while keeping your data organized within a central tool.
This platform offers push-button bioinformatics capabilities with publication quality analysis and reports. Learn more about the CosmosID-HUB’s capabilities.
Closing Thoughts
In summary, microbiome sequencing is a complex, yet highly innovative, tool available to microbiologists that opens the door to the microbiome world.
The unbiased, untargeted characterization of microorganisms within samples is unlocking untapped potential in various applications, such as nutritional research, live biotherapeutic drug prospects, environmental screening for pathogens (wastewater surveillance etc.), and so much more.
As the microbiome field continues to evolve, CosmosID will continue to be at the forefront of innovation and service for all microbiome researchers in their efforts, including yours!
Unlock the Power of the Microbiome with CosmosID…
No matter what part of the microbiome sequencing process you need assistance in, CosmosID has the experts and solutions to ensure that your research has what it needs to succeed. These solutions include:
Enquire today to speak to a member of our qualified team.
What is Microbiome Sequencing? FAQs
What are the steps in microbiome sequencing?
To properly perform microbiome sequencing, the 5 steps that must be accounted for include: Sample Collection, DNA Extraction, Library Preparation, Sequencing, and Data Analysis & Visualization.
What are sequencing techniques for the gut microbiome?
Multiple techniques can be used for human gut microbiome sequencing. Typical techniques are either amplicon sequencing (V4 region) or whole genome metagenomic shotgun sequencing (shallow or deep).
For whole genome shotgun sequencing, if research is interested only in strain-level taxonomic classification of organisms, shallow is sufficient. However, if functional and/or other gene detection is of interest, deep shotgun sequencing may be recommended.
What is metagenomic sequencing?
Metagenomic sequencing involves the process of sequencing the total DNA content of a sample, thus generating genetic signatures spanning the entire genome of all microorganisms within a sample.
In doing so, a significant amount of signatures (both taxonomic and functional) are captured, enabling a more accurate and robust characterization of the microbiome when compared to amplicon sequencing.
Want more like this? Sign-up to our newsletter to get the latest news from CosmosID: