Written by Nayeim Khan
The investigation of microbial communities and microbiomes has been revolutionized by the development of high-throughput sequencing. There are two main methods that most researchers use to analyze samples – sequencing all of the DNA in a sample (whole genome sequencing, WGS) or targeting a specific marker gene (amplicon sequencing). The premise of amplicon sequencing is based on identifying a gene or a marker that is present in most or all of the microbes you want to examine, but which also has enough variability from species to species to be able to discriminate between them. In 16S Amplicon sequencing a particular genetic locus, for example, the 16S rRNA gene in bacteria, is amplified from DNA extracted from the community of interest, and then sequenced on a next-generation sequencing platform. This technique removes the need to culture microbes in order to detect their presence and cost-effectively provides a deep census of a microbial community.
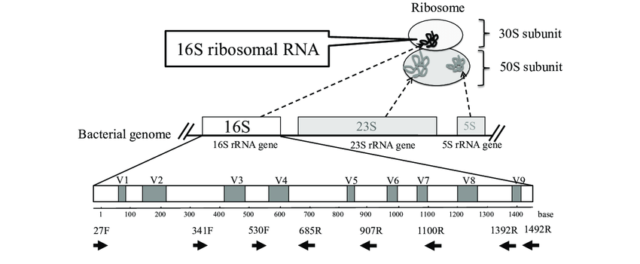
In order to aid scientists in quickly and effectively transforming their 16S amplicon data into actionable insights, CosmosID-HUB Microbiome is pleased to announce a new release of the 16S Amplicon workflow. The new 16S workflow uses a state-of-the-art denoising algorithm to model the errors introduced during amplicon sequencing. This is then used to infer true composition based on amplicon sequence variants (ASV), a finer resolution method than traditional operational taxonomic units (OTU). Besides the taxonomic classification, the new 16S workflow allows users to profile the predicted functional potential of the microbiome community using MetaCyc Pathways and Enzyme Commission enzymes definitions. The taxonomic and functional profiles generated using the 16S workflow can be aggregated on CosmosID-HUB Microbiome using associated metadata and compared across different cohorts using dynamic charts, graphs, statistical significance testing, and biomarker discovery analysis.
The new release of the 16S workflow is part of CosmosID’s microbial genomics platform, CosmosID-HUB Microbiome, which is continually expanding with various features and workflows for analyzing microbial communities and for inferring translational and actionable microbiome insights. Currently, CosmosID-HUB Microbiome analyzes 16S, whole genome, and ITS sequencing data and provides options for discovering antimicrobial resistance and virulence markers, microbial taxa (composition), and function. The overall CosmosID-HUB platform includes our COVID-19 pipeline, which detects the presence of COVID-19 and characterizes variants of concern or interest in both clinical and wastewater specimens. The HUB provides researchers with the flexibility to access and pay for the type of analysis they require.
To learn more about our 16S pipeline, please see our documentation here.

