Are you curious about the two main types of DNA sequencing technology? Nanopore and Illumina are both powerful tools in genetics research, but it can be difficult to understand how they differ.
In this guide, we’ll take a close look at both technologies and compare them side-by-side to help you make an informed decision when selecting the right sequencer for your project. We’ll examine their differences, advantages, and accuracy so that you can choose the best option for your specific needs.
With this guide as your starting point, you should have all the information necessary to make an educated choice between Illumina and Oxford Nanopore sequencing technologies.
What are Nanopore and Illumina Sequencing Platforms?
Nanopore and Illumina sequencing technologies are both powerful techniques used to sequence genetic material.
Nanopore is a single molecule-based technology that utilizes an enzyme to detect each nucleotide base as it passes through a tiny pore, while Illumina uses individual fluorescent molecules attached to the ends of strands of DNA in a process known as “clonal amplification” to detect sequences and sequence data.
Let’s take a closer look at each technology and discuss the key advantages of each.
What is Nanopore sequencing?
Oxford Nanopore technologies developed a new generation of DNA/RNA sequencing technology. The technology relies on traversing DNA/RNA sequences from a flow cell which contains tiny holes, “nanopores” embedding an electro-resistant membrane.
Nanopores in the flow cell have their electrodes connected to a sensor chip which quantifies electric current variation induced by different nucleotides through traversing the nanopore with membrane embedding.
The electric current variation is a characteristic that differentiates different types of nucleotides, such as A, T, G, and C. Algorithms then decode the electro current variation and translate it into DNA or RNA sequence.
(You can think of the current as water flowing through a pipe. When an object enters the pipe, the flow of water is disrupted, just as DNA disrupts the current as it passes through the nanopore.)
What are the advantages of Nanopore sequencing?
There are a number of advantages to using a Nanopore sequencing platform.
- Firstly, an ultra-long read sequence generated by Nanopore sequencing, where the top exceeds 4 Mb in length, promotes genomic assembly, as well as analysis of copy number and presence/absence variants, and repetitive regions.
- Nanopore sequencing is also the only sequencing technology that offers real-time analysis (for rapid insights), in fully scalable formats, can analyse native DNA or RNA, and can sequence any length of a fragment to achieve short to ultra-long read lengths.
- The portability of Nanopore sequencers promotes real-time analysis of data on the field, as well as projecting monitoring power to environments where otherwise genomic profiling may not be possible.
- The different designs of flow cells used in Nanopore sequencers also allow for greater scalability, due to the flexibility of this method.
What is Illumina sequencing?
Illumina sequencing technology leverages clonal array formation and proprietary reversible terminator technology for rapid and accurate large-scale sequencing. The innovative and flexible sequencing system enables a broad array of applications in genomics, transcriptomics, and epigenomics.
An Illumina sequencing platform uses sequencing by synthesis (SBS) technology relying on four fluorescently-labelled nucleotides to sequence the tens of millions of clusters on the flow cell surface in parallel.
During each sequencing cycle, a single labelled nucleotide is added to the nucleic acid chain.
The nucleotide label serves as a terminator for polymerization, so after each nucleotide incorporation, the fluorescent dye is imaged to identify the base and then enzymatically cleaved to allow incorporation of the next nucleotide.
Since all four reversible terminator-bound nucleotides (A, C, T, G) are present as single, separate molecules, natural competition minimises incorporation bias. Base calls are made directly from signal intensity measurements during each cycle, which greatly reduces raw error rates compared to other technologies.
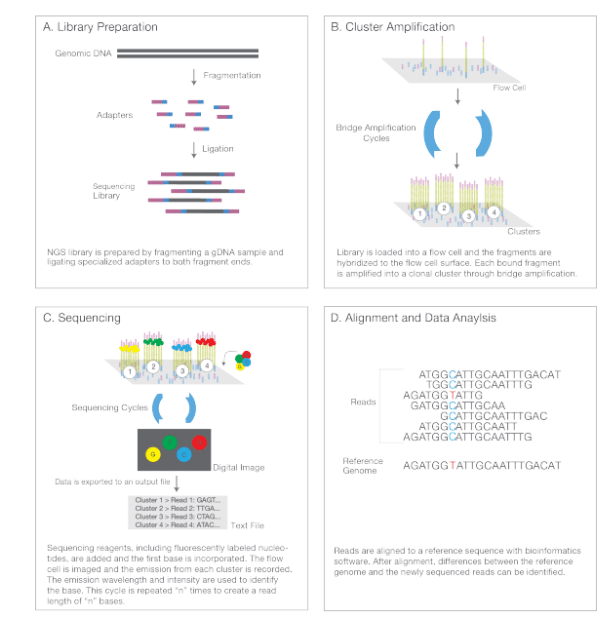
(Next-Generation Sequencing Chemistry Overview—Illumina NGS includes four steps: (A) library preparation, (B) cluster generation,(C) sequencing, and (D) alignment and data analysis.)
What are the advantages of Illumina sequencing?
Illumina data sequencing also offers a number of benefits:
- Illumina sequencing results are highly accurate base-by-base sequencing that eliminates sequence-context specific errors, enabling robust base calling across the genome, including repetitive sequence regions and within homopolymers.
- The high accuracy of Illumina sequencing also enables a wide variety of applications, allowing researchers to ask virtually any question related to the genome, transcriptome, or epigenome of any organism, making it a fit method in clinical settings.
Illumina vs Nanopore Sequencing: What are the similarities?
Both Illumina and Nanopore sequencing are high-throughput sequencing methods, meaning they can sequence many DNA templates in parallel, providing researchers with reliable data in rapid timeframes.
Both methods are suitable for whole genome sequencing, from small to large organisms, with very different sequencing read lengths depending on the technology used. Thanks to both methods, we can sequence DNA content from isolate or bulk samples to construct an isolate genome or transcriptome, as well as a metagenome or metatranscriptome.
Additionally, by creating a whole genome sequence from a sample using either technology, we enable a wide variety of applications in genomics, transcriptomics, and epigenomics.
Nanopore Sequencing vs Illumina: What’s the difference?
Next-generation sequencing (NGS) methods differ primarily by how the DNA or RNA sequencing method is prepared and the data analysis options used.
Nanopore sequencing generates long reads of DNA sequence quantifying variable electromagnetic field variation induced by different nucleotides. Conversely, an Illumina sequencer uses dye terminators that quantify short reads of DNA sequences by variable wavelength emission of dye colours.
Below, we’ve split the differences between the two technologies into three key categories: sequencing method, read length and strengths.
Difference 1: Sequencing method
One main difference is that both sequencing platforms use different sequencing methods.
Oxford Nanopore technologies use the variation in a nanopore’s membrane current to detect differences between nucleotides, while Illumina sequencing utilizes dye terminators bound to the four bases (A, C, T, G) and measures the variable wavelength emission of each dye color.
Difference 2: Length of reads
Nanopore sequencing can produce long reads of up to hundreds of kilobases, while Illumina sequencing is limited to short read lengths of up to 500 bp.
The maximum read length for Nanopore sequencing is dependent on the sample quality and target length, but in general, Nanopore sequencing produces reads that are longer than Illumina sequencing data.
Difference 3: Strengths
In short: nanopore is better at assembly and portability, and Illumina at accuracy.
On one hand, the accurate short reads of Illumina sequencing make it fit for clinical samples but may complicate genome assembling. On the other hand, the long-read sequencing of Nanopore sequencing data, with ease for genome assembling, is less accurate than short sequencing reads of Illumina sequencing.
Additionally, Nanopore sequencing is portable and allows real-time analysis in remote regions, while Illumina sequencing requires a dedicated lab setting.
Nanopore Sequencing or Illumina: Which is best?
Choosing the best NGS technology for a project requires careful consideration, weighing up the needs of the research or experiment in order to select an appropriate tool.
Overall, both methods are powerful tools for conducting genome and transcriptome analysis, but depending on the application it may be more suitable to use one method over the other.
For example, Illumina would be the better option for clinical diagnostics, or other settings with high accuracy demands, whereas Nanopore sequencing may be a better option for field projects, as well as analysis for CNV and repetitive regions.
It is also important to note that both Illumina and Nanopore sequencing can be used in combination, leading to the most comprehensive insight into genomic data.
In any case, a better understanding of why and when each method should be used is key for unlocking the most valuable insights from your data. With a clear understanding of both Nanopore and Illumina sequencing, scientists are able to make an informed decision about which is best for their particular projects.
CosmosID Sequencing Services
At CosmosID, we provide genome sequencing services and bioinformatic analysis for both Nanopore and Illumina sequencing, enabling researchers to make use of the best method for their specific needs.
With our expertise in genomics, we can help you select the right sequencing technology, as well as provide a comprehensive analysis of your data. Contact us today to learn more about how our CosmosID-HUB can help you drive your research forward.
Interested in sequencing? CosmosID could help you with shallow and deep Illumina shotgun metagenomics and metatranscriptomics sequencing.
Want more like this? Sign-up to our newsletter to get the latest news from CosmosID: